Beamline citations and MAX IV acknowledgements
Please cite the following papers when publishing results based on experiments at NanoMAX. Also note that MAX IV requires acknowledgement of beamtime, see the policy pages for the formal statement.
- Björling et al., Optics Express (2020), for the coherent nanofocused beam.
- Johansson et al., Journal of Synchrotron Radiation (2021), for the beamline instrumentation and performance.
- Carbone et al., Journal of Synchrotron Radiation (2022), for the design of the diffraction endstation.
Experimental data download
Data can be downloaded either using Globus, or via normal sftp. The following page contains details and instructions on how to proceed.
https://www.maxiv.lu.se/beamlines-accelerators/controls-it/it-services/
Data format
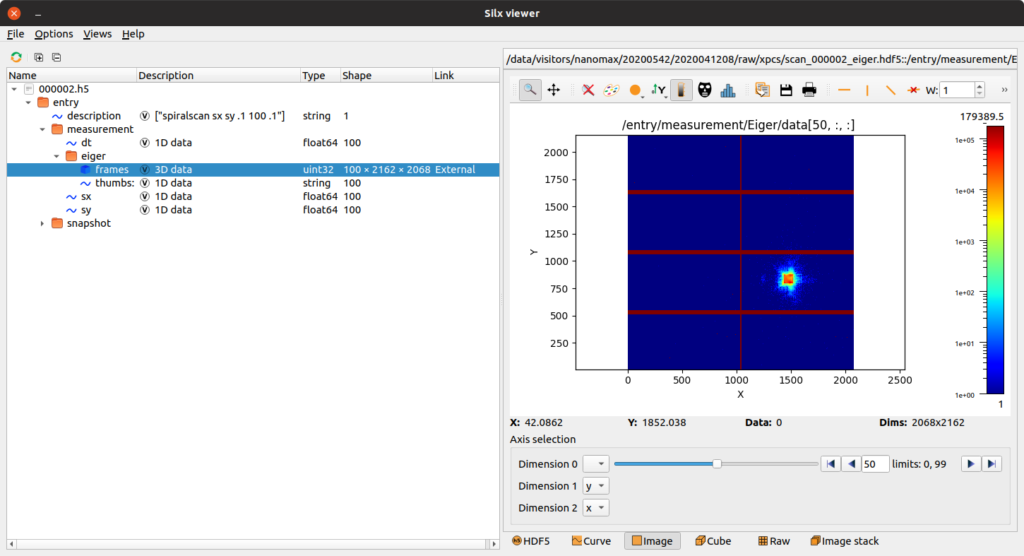
NanoMAX records data as compressed hdf5 files, a binary file type which stores data in a tree-like hierarchy. The structure of data collected at NanoMAX is rather simple, with one main file collected per scan performed. Using freely available viewing tools, these files can be viewed and explored. While heavy data (like 2D images and XRF spectra) are stored in separate detector files to allow more efficient data acquisition, the main file links to these data and provides a unified way of accessing the data, provided that the detector files can be found in the same folder. In this example, a scan across 100 positions on the motors sx and sy were recorded, providing 100 motor positions and 100 detector images. The main file also contains the scan command used, as well as a snapshot of the beamline at the start of the scan.
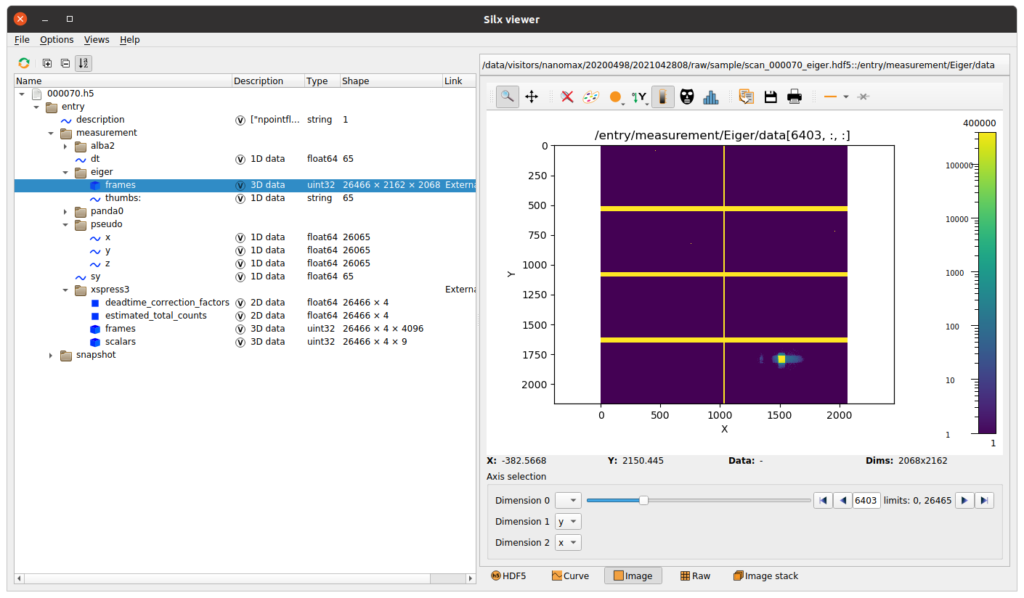
A more elaborate dataset, collected by continuous scanning of the piezo stage, is shown below. Here, the positional information is contained in the arrays pseudo/x and pseudo/y, while a collection of detectors, like the eiger pixel detector and xspress3 fluorescence detector were simultaneously used.
Decompressing Eiger and Pilatus data
These detectors write data compressed with the bitshuffle algorithm. There are various ways of installing the required decompression filters, but the recommended way is to work within a conda environment, for which the MAX IV conda channel provides the necessary Linux packages. Data can then be browsed as above and visualized as below.
conda config --add channels maxiv conda install bitshuffle
For windows and mac, use the hdf5plugin module. The most convenient way is to use Anaconda, in which the following command installs the necessary package.
conda install -c conda-forge hdf5plugin
Basic experimental data visualization
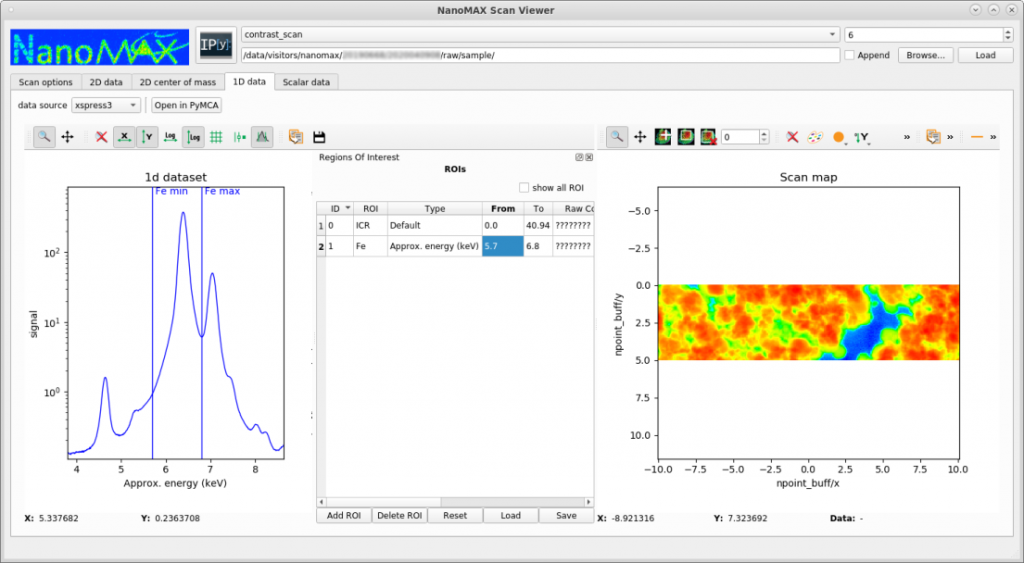
NanoMAX provides a basic tool for quickly visualizing scanning data, such as XRF or diffraction mapping. The tool is not designed for quantitative fitting, but for generating fast overviews and evalutating ongoing experiments. The tool (called scanViewer) can be downloaded here. It is only developed and tested on Linux, but users have reported using it successfully with Mac and Windows. In the following example, a region of interest of a fluorsscence emission spectrum is selected, and the intensity in this region is mapped out across the sample.
Fluorescence image data analysis
We normally use the software PyMCA developed at ESRF for fluorescence imaging data analysis. XRF image analysis can be a bit tricky in the beginning and therefore some assistance can be found in this step-by-step guide describing how to load and fit NanoMAX X-ray fluorescence data with PyMCA.