Imagine this future. Vehicles and machinery primarily powered by renewable organic matter, a resource far better for the planet’s health than today’s predominate fossil fuels. What factors stand in the way for a global power transition to competitive, industrial-scale biomass conversion? A study in Nature Communications reveals one key piece of the puzzle using bacterial enzymes. At MAX IV’s BioMAX beamline, an international team of scientists has determined important rate-limiting steps of lignocellulose breakdown, a major hurdle in efficient biomass processing. The discovery holds promise for a significant reduction in manufacturing costs and faster adoption of new biomass-derived fuels to market.
“Our results could be very useful to guide how much enzyme is needed for a biomass degradation process or determine what aspects of an enzyme to try to modify and what substrates are relevant to test activity on, where small model compounds do not properly mimic the more complex cell wall,” said Johan Larsbrink, study researcher and associate professor of molecular enzymology at Chalmers University of Technology in Sweden. “If more complex and larger substrates could be generated and become more widely available, these would enable, for example, enzyme engineering in a more streamlined fashion.”
The majority of lignin conversion studies in the past have focused on degradation of biomass by white or brown fungi, both recognized lignin degraders in nature. Garnering interest is a newer area of lignin research utilizing bacteria for its decomposition ability, metabolic versatility, and potential use in the synthesis of lignin-derived chemicals for biotechnology.

The researchers’ curiosity for bacterial enzymatic action stemmed from several studies characterizing the breakdown of plant cell walls with fungal enzymes. The referenced enzyme family contained many unstudied bacterial sequences, and the known enzymes performed unique enzymatic functions for degradation. Larsbrink and colleagues’ succeeding investigation went further along this path, describing a selection of ten bacterial enzymes much more diverse in both sequence and structure compared to fungal enzymes and with broader substrate specificity. The chain of knowledge led to their most recent revelation: identification of rate-limiting steps of a bacterial enzyme.
Breaking down cell walls
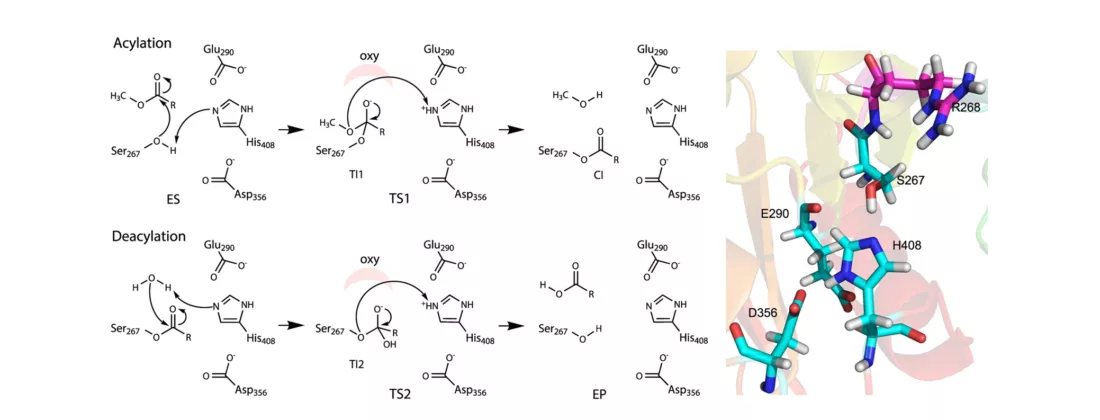
The researchers investigated the catalytic machinery of biomass conversion using bacterial enzyme, glucuronoyl esterase (GE) OtCE15A on a highly complex substrate, lignin carbohydrate complex (LCC). With measurements at BioMAX beamline, the step of deacylation, which removes an acyl group, was determined to be rate-limiting as it significantly influenced the breakdown of LCC. The GE enzyme weakens the robust molecular linkage between lignin—the binding polymer that provides strength to plant cell walls—and the cell wall polysaccharides. In addition, the group’s calculations revealed the non-catalytic event of dissociation could be the rate-limiting step for large substrates often used in industrial enzyme engineering.
“The [enzymatic] interaction to complex biomass is a very important aspect of catalysis that is difficult to probe using regular substrates,” explained Larsbrink. “The dual catalytic acid of this particular enzyme we studied is interesting and still a bit puzzling as most other homologous enzymes have only one catalytic acid residue in one of the two equivalent positions, where the dual setup is found in OtCE15A.”
The research group is currently continuing the work on structure-function studies of glucuronoyl esterases and similar enzymes, funded by the Novo Nordisk Foundation, and will compare new and previously studied enzymes or their genomic context. Larbrink’s group at Chalmers is also looking at other esterases involved in biomass conversion such as tannases, and microbial degradation of other types of biomass apart from wood and agricultural waste.