The following is a summary of the IT facilities for BioMAX users. Click on each of the titles below to expand the contents.
BioMAX has several Linux workstations running the GNOME desktop dedicated for data collection. The naming convention for these workstations is b-biomax-controlroom-c-‘#’. There is also a dedicated machine for remote experiments, biomax-remote.
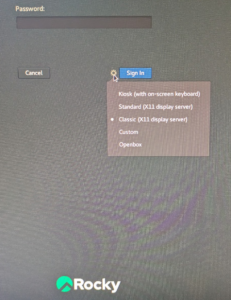
Since July 2023 all the beamline workstations are running Rocky Linux. On the beamline computers, we recommend changing the default window manager setting to the Classic X11 display server at the password prompt window, after you type your user name. Before typing the password, use the settings wheel to the left of the blue Sign in button and select the Classic option. The machine will remember your choice for future sessions. Remote users do not need to do this, since the biomax-remote server automatically forces the Thinlinc remote desktop to use the classic window manager.

The standard desktop on all the beamline machine contains several icons that fast-link to useful applications for the experiment, like the Albula monitor or ADXV to view diffraction images, links to MXCuBE, EXI and the Beam Position monitor tool.
Note: The very first time you log in to a beamline workstation or the remote server you will not see any desktop icons to MXCuBE, EXI and other software. You have to log out and log in again to see them.
Other software can be accessed from the Applications menu on the Desktop top bar (classic display only).
All the beamline computers as well as the biomax-remote server are located in the MAX IV blue network. The blue network is rather isolated to protect the beamline control software that runs on it, and in general, it is not possible to access other computers outside this network from it (the only exception is the online cluster for data processing, see next section) or link to web sites outside MAX IV.
There are two HPC clusters available to all MAX IV users:
- The online cluster, dedicated to data analysis during the beamtime. The front end machine are clu0-fe-2 and clu0-fe-3 (clu0-fe-1 has been deprecated). The online cluster can be accessed from the beamline workstations or biomax-remote via the Thinlinc desktop client, by clicking the Applications menu on the top panel and selecting the Internet submenu.
- The offline cluster, a smaller cluster that can be used to reprocess data manually after or during the beamtime. The front end is called offline-fe1. The offline cluster is not accessible from the beamline computers or biomax-remote, but it is possible to log in to it remotely from the users’ home computer. Please refer to these instructions.
In July 2024 the HPC clusters were updated to Rocky Linux 8.1. The front end Thinlinc servers use Xfce as the window manager. See this video for an introduction to the new look.
Unlike the beamline computers, The HPC clusters are on the MAX IV white (office) network and it is possible to connect to external sites from them.
Important: The HPC clusters are shared resources. There are limits to the maximum number of users who can log in at one time. Therefore, please log out always when you have finished working. There is also a limit to the available processors. Although the data processing needs of crystallographic experiments are relatively modest, for special projects that require many hours of computation and several nodes on a regular basis, please consider requesting access to LUNARC.
See also Data handling and processing at BioMAX and HPC basics .
All the MAX IV computers available to the user mount the /data disk and the /mxn disk. /mxn/visitors/'yourusername' is the home directory of the beamline workstations. Note, however, that the home directory in the HPC clusters (/home/'yourusername') is different and not mounted on other machines, and it will not be visible from the beamline control computers or, when transferring files, the sftp server.
All the raw data and automated processing results are stored in subdirectories under /data/visitors/biomax or /data/proprietary/biomax for proprietary proposals.
The home directory can be used to store files related to the experiments, such as PDB files and processing scripts. They should not be used to store data images under any circumstances. Both the home directory under /mxn/visitors and /home (in the clusters) are backed up and up to four images are saved. To access snapshots of a directory less than one week old, go to the subdirectory .snapshot (this directory is not be visible until one accesses it).
We recommend that users transfer the data from home using either Globus or sftp. Backing up data locally to an external hard disk is no longer allowed to reduce the risk of malware infection.
Public data sets can be downloaded from SciCat.
See also the Guide to IT services.
If the videos looks blurry, change the “Quality” setting (under the cogwheel icon below the video) to 1080p.